| News / Tech News |
Identifying proteins using nanopores and supercomputers
The amount and type of proteins human cells produce provide important details about a person's health and how the human body works. But the methods of identifying and quantifying individual proteins are inadequate to the task, scientists say. Not only is the diversity of proteins unknown, but often amino acids are changed after synthesis through post-translational modifications.
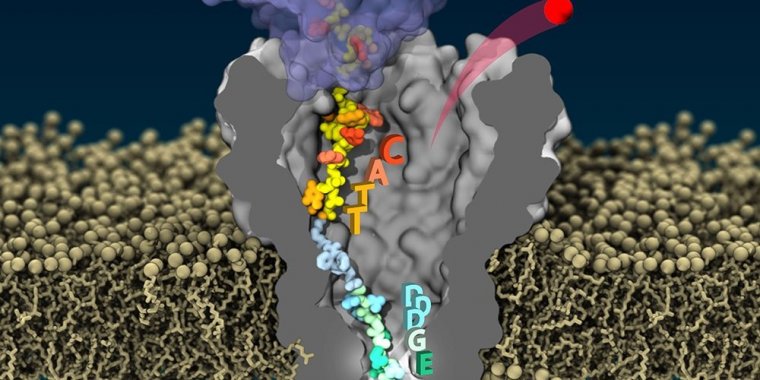
High-fidelity reading of single protein composition by pulling the same protein through a nanopore. Photo: Jingqian Liu
Researchers at Delft University of Technology in the Netherlands and the University of Illinois at Urbana-Champaign have expanded on DNA nanopore successes and provided a proof-of-concept that the same method is possible for single protein identification, characterizing proteins with single-amino acid resolution and within a small margin of error.
The workhorses of our cells, proteins are long peptide strings made up of 20 different types of amino acids.
The researchers used an enzyme called helicase Hel308 that can attach to DNA-peptide hybrids and pull them, in a controlled way, through a biological nanopore known as MspA.
Lead author Henry Brinkerhoff likens the protein to a necklace with different-sized beads. "Imagine you turn on the tap as you slowly move that necklace down the drain, which in this case is the nanopore," he said.
"If a big bead is blocking the drain, the water flowing through will only be a trickle; if you have smaller beads in the necklace right at the drain, more water can flow through."
By loading a liquid medium with helicases, the researchers can get many separate, overlapping reads of the same molecule; they can "rewind" the protein and read its amino acid sequence again. Doing so reduced errors from 13% to practically zero. (National Science Foundation)





