| Health / Health News |
New prediction algorithm identifies previously undetected cancer driver genes
A new study, led by researchers at the University of California, Irvine, has deepened the understanding of epigenetic mechanisms in tumorigenesis and revealed a previously undetected repertoire of cancer driver genes.
New algorithm reveals undetected cancer driver genes. Photo: UCI School of Medicine
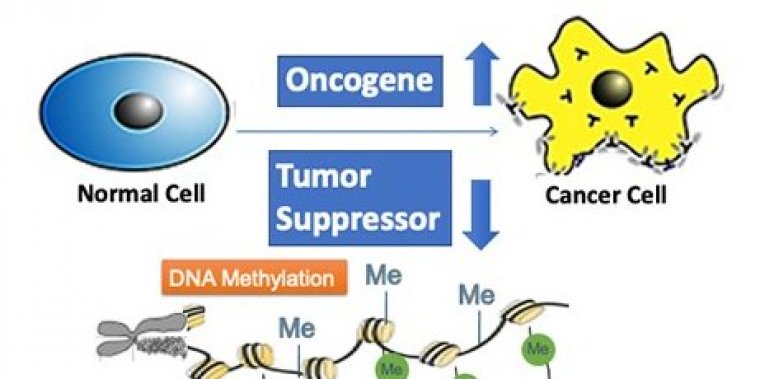
Using a new prediction algorithm, called DORGE (Discovery of Oncogenes and tumor suppressoR genes using Genetic and Epigenetic features), researchers were able to identify novel tumor suppressor genes (TSGs) and oncogenes (OGs), particularly those with rare mutations, by integrating the most comprehensive collection of genetic and epigenetic data.
"Existing bioinformatic algorithms do not sufficiently leverage epigenetic features to predict cancer driver genes, despite the fact that epigenetic alterations are known to be associated with cancer driver genes," said Wei Li, senior author of the study. "Our computational algorithm integrates public data on epigenetic and genetic alternations to improve the prediction of cancer driver genes."
Cancer results from an accumulation of key genetic alterations that disrupt the balance between cell division and apoptosis. Genes with "driver" mutations that affect cancer progression are known as cancer driver genes and can be classified as TSGs and oncogenes OGs based on their roles in cancer progression.
This study demonstrated that cancer driver genes, predicted by DORGE, included both known cancer driver genes and novel driver genes not reported in current literature.
In addition, researchers found that the novel dual-functional genes, which DORGE predicted as both TSGs and OGs, are highly enriched at hubs in protein-protein interaction and drug/compound-gene networks.
"Our DORGE algorithm successfully leveraged public data to discover the genetic and epigenetic alterations that play significant roles in cancer driver gene dysregulation," explained Li. "These findings could be instrumental in improving cancer prevention, diagnosis and treatment efforts in the future." (National Science Foundation)
YOU MAY ALSO LIKE





