| News / Science News |
Study shows limitations of method for determining protein structure
A new study by chemists at the University of Arkansas shows that X-ray crystallography, the standard method for determining the structure of proteins, may provide inaccurate information about a critical set of proteins -- those found in cell membranes -- which in turn could be leading to poor and inefficient drug design.
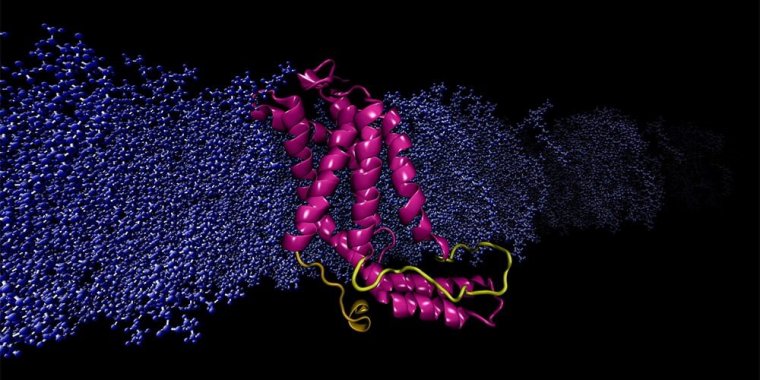
Results of a new study of protein structure may lead to better drug design. Photo: Sogol Moradi
"Two-thirds of all drugs, including those used for chemotherapy, target proteins found on cell membranes," said Mahmoud Moradi, a biochemist at the university. "Unfortunately, X-ray crystallography, the gold standard for determining the structure of proteins, has many limitations when dealing with those found in the cellular membrane. Our work exposes, and in many ways, explains these limitations."
Considered the workhorse molecules of cells, proteins are responsible for nearly every task in living systems. Some proteins live inside the cell, and some reside on the cell's membrane, an outer layer of lipids that separates the cell from its external environment.
Membrane proteins are critically important: They regulate the exchange of information and materials between the cell and its environment, a vital task for survival and normal function because any disorder in protein function can result in disease.
The study of protein function is needed to understand the molecular basis of disease. Moradi and his team have addressed the problem of relying on X-ray crystallography by using a supercomputer at the Arkansas High Performance Computing Center to run continuous, microsecond-level computations simulating the molecular dynamics of membrane proteins.
The results demonstrate that computational chemistry and supercomputing technology can be used to model membrane proteins more accurately in an environment that mimics their physiological environment. (National Science Foundation)
YOU MAY ALSO LIKE





